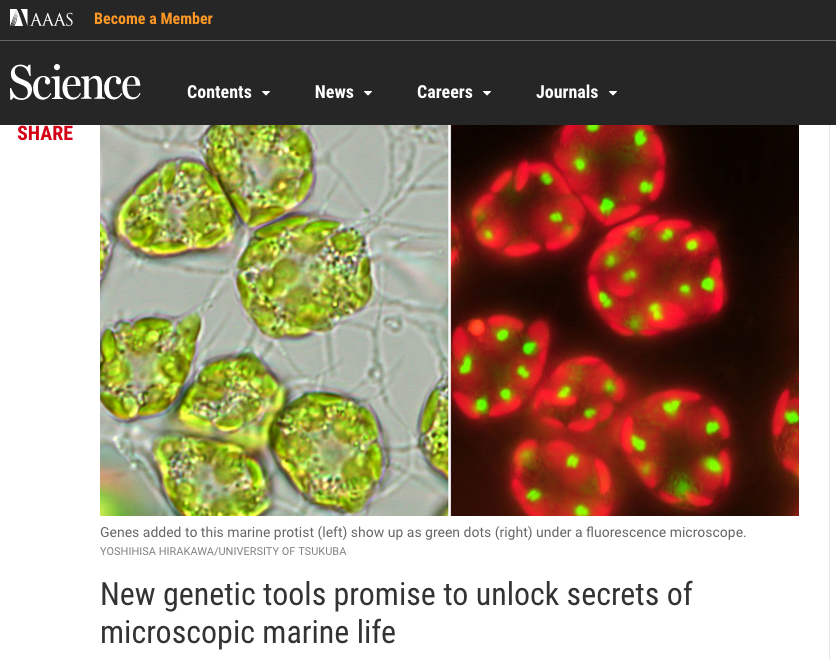
MOCK RESEARCH LAB
School of Environmental Sciences, University of East Anglia, Norwich Research Park, Norwich, UK
Towards digital plankton twins of the ocean. 5x preprints just out on *Zenodo*
New collection at *Scientific Data* open for submissions: Biological Diversity and Environmental Change
Mission Statement
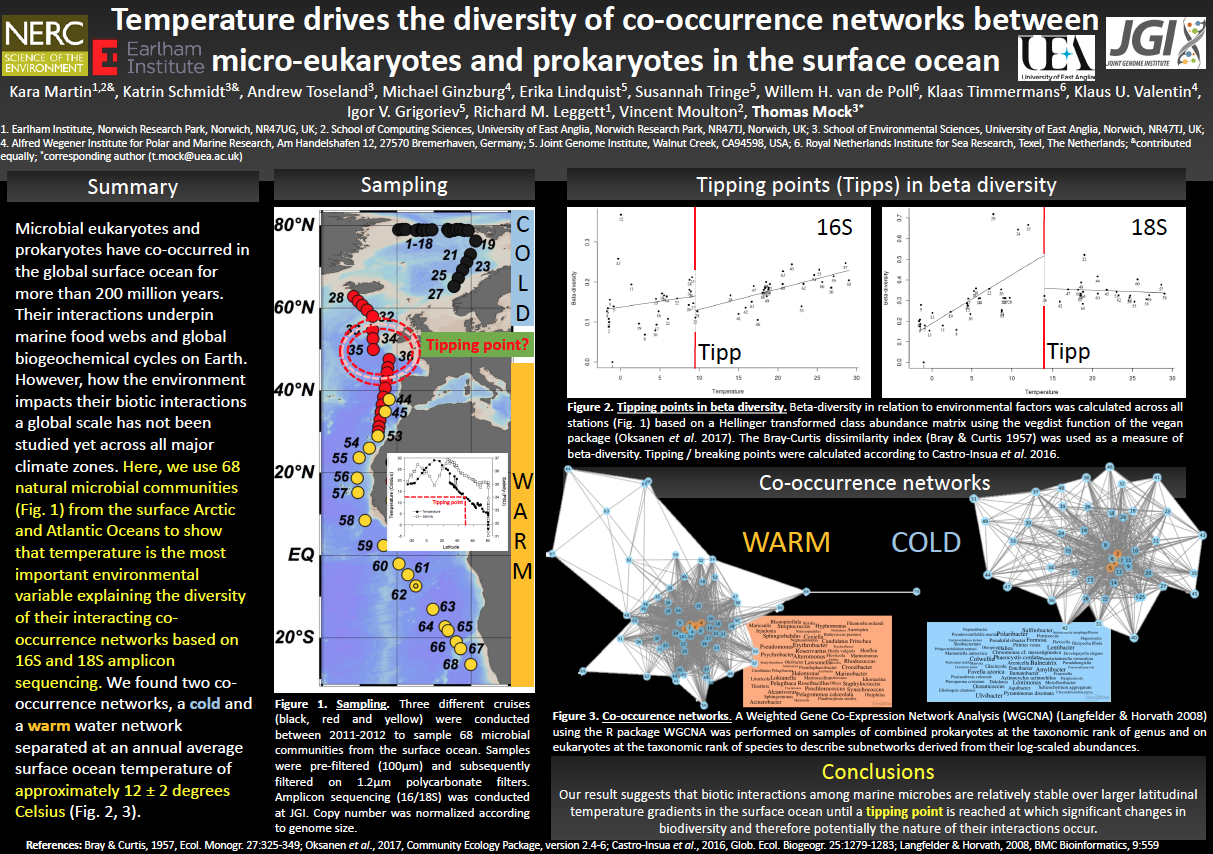
The overarching aim of our research is to identify fundamental biological processes that govern the adaptation and evolution of marine microalgae (Phytoplankton) and their microbiomes (e.g. bacteria) in the oceans. We want to know how things work from small to large scale. To meet this aim, we integrate approaches from cell biology, reverse genetics, physiology, multi-omics and ecology. We focus on marine algae and their microbiomes because they contribute ca. 50% of annual global carbon fixation and therefore significantly contribute to the habitability of our Earth. Thus, their adaptation and (co)evolution with associated microbes (e.g. bacteria and viruses) in the context of biotic and abiotic (e.g. temperature, nutrients) interactions in different marine systems (from tropics to the poles) and under conditions of climate change does not only shape the nature of marine food webs but also biogeochemical cycles they drive. This fundamental knowledge will help us to improve predictions on how algal microbiomes respond to global climate change.
Overview
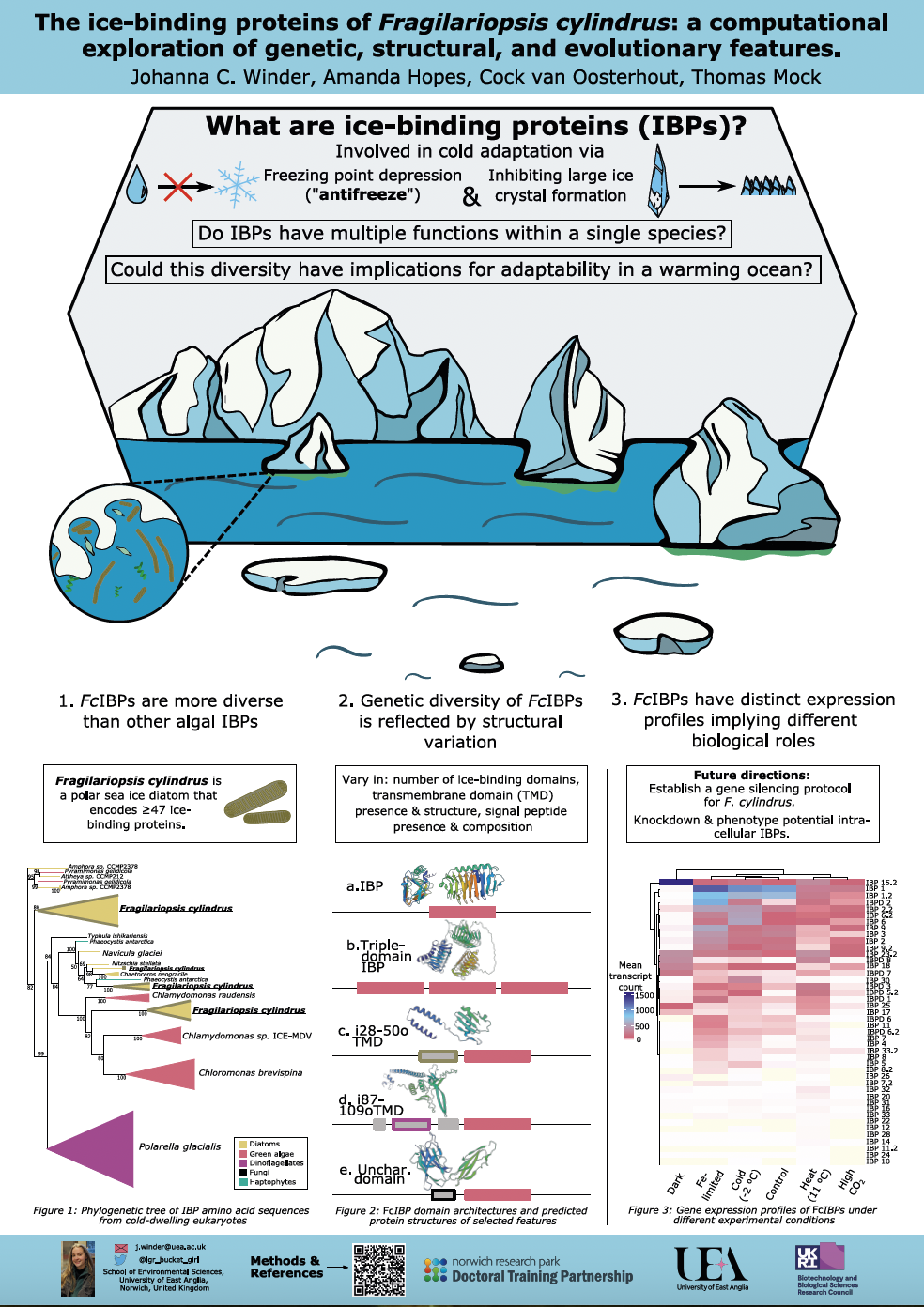
Our group mainly works in the fields of evolution, cell biology, reverse genetics and genomics (e.g., PACBIO and Oxford Nanopore sequencing; CRISPR/Cas) with marine algae (e.g., Thalassiosira pseudonana, Fragilariopsis cylindrus - diatoms) and their microbiomes (e.g., prokaryotes, viruses) from the global upper ocean to understand their ecology, (co-)evolution and adaptation. This work leads to the identification of biological mechanisms that shape their evolution and are therefore responsible for their unique biology and adaptation to different environmental conditions including climate change.